Unique cancer discovery opens path to early diagnosis and specialised treatment breakthroughs
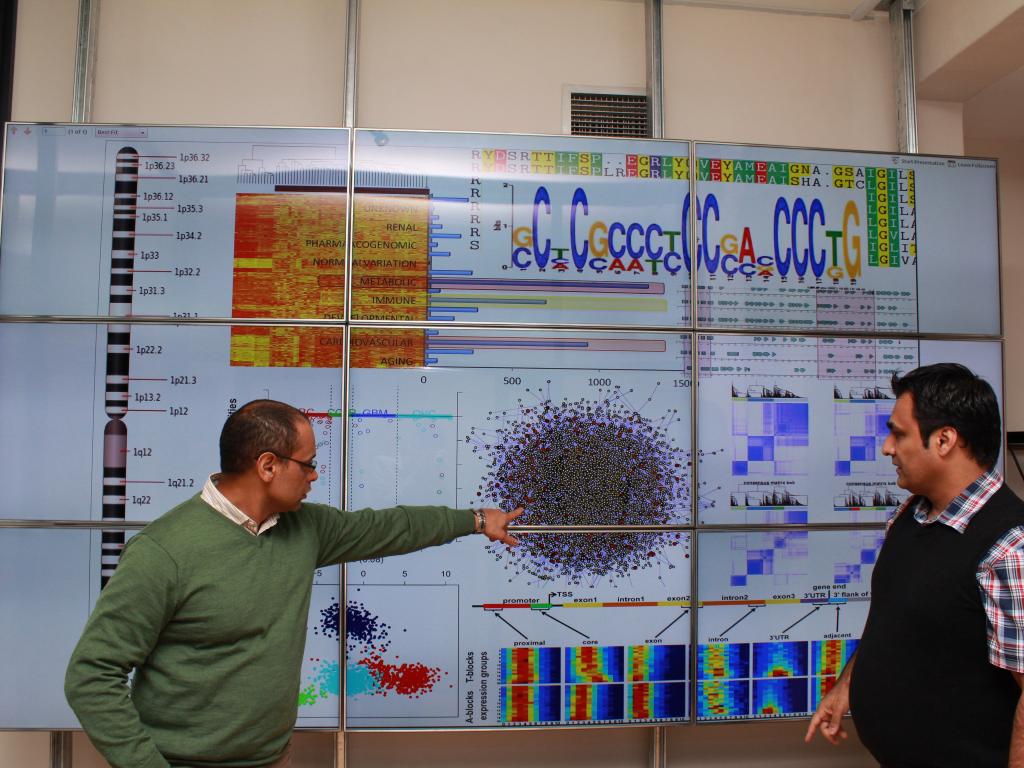
Professor Kevin Naidoo and Dr Jahanshah Ashkani with a model of the GT gene profiles
Each of six cancer types (breast, colon, lung, kidney, ovarian and brain) has a unique genetic expression pattern that can be used for accurate early diagnosis and targeted treatment, a team based at the University of Cape Town has discovered.
Using statistical classification algorithms on massive tumour gene expression data, the researchers found that the GT gene expression pattern of a patient can be used to accurately classify cancer types that lay the ground for developing an early diagnostic. Equally impressive is that the expression patterns can identify variations within each of the cancer type, which can then guide specialised patient treatment.
Professor Kevin Naidoo, the SA Research Chair in Scientific Computing in the Department of Chemistry at UCT, and Dr Jahanshah Ashkani, also of the UCT Chemistry Department, made the discovery. Professor Naidoo is now leading a multi-laboratory collaboration that includes scientists in the divisions of pathology and human genetics at UCT’s medical campus and the Centre for Proteomics and Genomics Research to analyse the blood samples of South African patients. They hope to develop a low-cost gene expression tool for breast cancer that will form the basis of a routinely used early diagnostic.
An early cancer diagnostic is critical for patient survival, as most cancers can be cured if discovered in their early stages. Of similar importance is the choice of treatment once a patient has been diagnosed with cancer. The use of the GT gene profile method of identifying the distinct subtypes of breast and other cancers opens the door to further research that will guide the choice of specialised treatment to significantly enhance a patient’s chances. This dovetails with the shift towards personalised medicine approaches that deliver specialised oncotherapy to patients following diagnosis.
Singling out 210 gene types from cancer using big data
This discovery demonstrates the importance of computational big data analytics in biomedical sciences and the developing field of personalised medicine.
The discovery of a cancer-type carbohydrate related gene signature led the team to a genomic classification of cancer types. Their work, published in the current issue of Scientific Reports, describes the statistical analysis of 1893 patient tumour gene expression data from The Cancer Genome Atlas (TCGA). This analysis was made possible through the use of computational big data analytics – the examination of large data sets containing a variety of data types to reveal, in this case, the hidden expression patterns of each cancer type.
The team identified GT gene profiles that regulate the enzymes (GTs) responsible for modifying the structures of complex carbohydrates coating the surfaces of tumour cells. By extracting and analysing only 210 GT genes from each of the 1893 full tumour genome sets (each displaying the expressions of more than 20,000 genes) the team found patterns in the data that led to their discovery of a gene expression classification of cancer types.
