ProtoCaller: Robust Automation of Binding Free Energy Calculations
SCRU, in collaboration with researchers from the UK (from universities including the University of Southampton and Cambridge) have developed a Python library which automates relative protein−ligand binding free energy calculations in GROMACS. These free energy calculations are an increasingly promising approach for facilitating drug discovery.
The setup of free energy calculations is a potentially more time consuming task than data generation. This is due to the multiple steps in the setup process of a simulation, each requiring some level of user intervention. A prominent limitation of the ease of use and efficiency has been software interoperability. The necessary linking of different softwares increases the risk of human error when trying to interface the software with in-house scripts.
ProtoCaller solves these problems by providing a customizable unified interface to all of the steps of the free energy workflow. ProtoCaller is also very flexible, being able to be tailored to the user’s needs, even if those were to extend to other calculations ( for example, absolute and relative solvation free energies and simulations in MD engines supported by BioSimSpace).
For more information, see ProtoCaller: Robust Automation of Binding Free Energy Calculations, Journal of Chemical Information Modelling 60, 1917 - 1921. https://pubs.acs.org/doi/10.1021/acs.jcim.9b01158
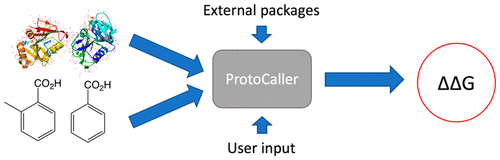
Fig. 1: Illustration of Protocaller’s effect on a workflow
