BRIDGE: An Open Platform for Reproducible Protein-Ligand Simulations and Free Energy of Binding Calculations
One of the main problems in predicting protein-ligand binding is the complexity of performing reproducible protein conformational analysis and ligand binding calculations, using vetted methods and protocols. Protein-ligand binding prediction is central to the drug-discovery process.
Upon reading the paper reporting the open-source BRIDGE platform (https://academic.oup.com/bioinformatics/article-abstract/35/18/3508/5317160?redirectedFrom=fulltext ) the editors of Bio Protocol, a journal that aims to make scientific discoveries open and reproducible ((https://bio-protocol.org/Default.aspx) invited Kevin Naidoo to write a paper describing the protocol for free energy computations. The SCRU Lab researchers wrote the article with the aim of sharing protocols seamlessly to enable more effective drug discovery calculations following genomics and proteomics discovery of protein drug targets. The authors show the dynamics and simulation protocols for predicting protein-ligand binding affinities in silico on the T4 lysozyme system. This was done by way of introduction to the BRIDGE platform which make available biomolecular simulations and methods available to novices and reproducible for validation by experts.
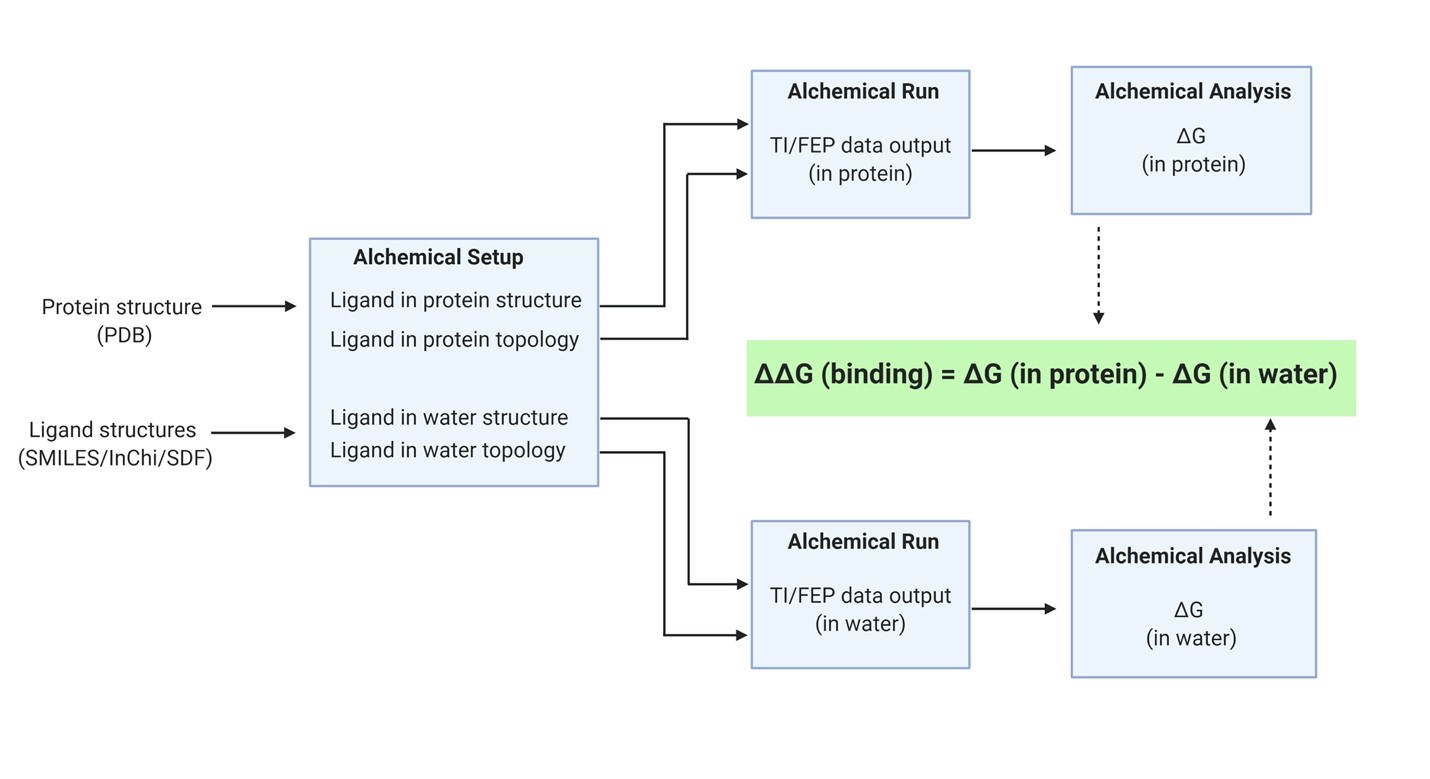
For more information see BRIDGE: An Open Platform for Reproducible Protein-Ligand Simulations and Free Energy of Binding Calculations in Bio-Protocol 10(17): e3731. https://bio-protocol.org/e3731
